Introduction¶
evolearn is a neuroevolution framework for designing artificial life experiments using variants of the HyperNEAT algorithm.
This manual is intended to be an introduction to the evolearn package, as well as its design and contribution to the fields of neuroevolution, artificial intelligence, and theoretical neuroscience according to the research goals of the author. You will find code for interacting with the library and using it to implement standard machine learning techniques and less common neuroevolutionary methods on a variety of tasks.
Once you have made your way through the Introduction, you can find a collection of experiments relevant to your neuroevolution research.
Overview¶
The goal of evolearn parellels the ideal (though often unspoken) goals of artificial intelligence - to develop solid theory for generating intelligent models for solving any type of problem a biological nervous system could encounter. That goal is tremenously ambitious, and in a way the purpose of evolearn is to expose the difficulty of striving towards that objective. Furthermore, evolearn focuses on neuroevolutionary methods of indirect encodings, taking inspiration from the mechanisms that allowed biological organisms to discovery meaningful and effective solutions for life through evolution by natural selection.
Biological organisms are intelligent in ways we hope to replicate in machines. There is a tremendous diversity of solutions of intelligence on our planet, and a theory of nervous systems should account for that diversity. A prominent way of thinking about the existence of a particular nervous system is the observation that it exists as a solution that is a product of both learning during the lifetime of that organism, as well as the evolutionary history of previous solutions that led to it.
It may not always be necessary to include evolutionary search in training models to solve certain classes of problems. Benchmark machine learning models in image recognition do not at this point include global search methods prior to incremental error propagation. In fact, it may be possible to find sufficiently good solutions without employing evolutionary computation at all. On the other hand, there are a few reasons for thinking that evolutionary thinking could improve the performance of some of these models, as well as the discovery of better ones.
First, it seems that the problem of escaping local minima when comparing solution performance against an objective is an inescapable problem. Local gradient-based changes to a model require specific alterations to avoid this issue. Global optimization approaches like genetic algorithms, however, can often discover multiple minima in the objective function and avoid getting stuck.
Second, because local gradient-based methods often require techniques to prevent them from getting bogged down in local minima, successfully implenting a class of models on a new dataset often requires significant expertise. If instead we can initialize simple models for a task and evolve them over time, the level of expertise needed to use them may be decreased.
Third, it is an often unspoken objective in the fields of machine learning and artificial intelligence themselves to search for models that can match and/or outperform the capabilities of biological nervous systems. While it may not be necessary to undergo billions of years of evolution to reach certain solutions in the space of all possible ones, it is a fact that in biology those solutions were reached through exactly that process. Therefore, it is at least worth our attention, when attempting to connect the performance of machine learning models to that of the organisms we study in neuroscience, that global search strategies could very well be responsible for both the level and diversity of intelligent life we see around us.
Getting Started¶
Let’s say for a moment that it was our goal to find an organism that can effectively move through an evironment and eat enough food to stay alive. Within that goal there are a few important components that can get us started towards modelling an abstract biological organism.
- The organism must move: we would like to create a model with outputs.
- The organism must consume food: this would imply there is feedback in our environment, such as a reward for nutrients.
- The organism must stay alive: this organism can learn and improve against this objective.
Since this package will completely revolve around artificial neural networks, it should come as no surprise that we will end up looking for solutions (organisms) that can be represented as a neural network. An organism can observe its environment, make observations about the food it can see, make actions in response to that input, and then receive reward feedback that can be used to alter the structure of that neural network so that it can be better in the future.
Ultimately we will be able to use evolearn on more complex agent-environment controllers, and even simulate controllers that can then be embodied in robotic agents in the real world - but, at least our agent can act as a starting point.
A Simple Controller Task¶
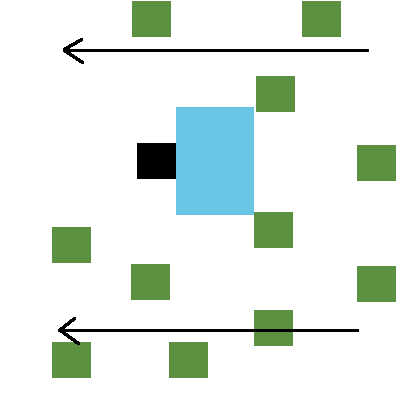
We can design our agent in an environment that resembles the popular reinforcement learning benchmark game Flappy Bird. (a single frame from this task is shown below in Figure 1)
An agent remains stationary in a column of a grid world, while the world continues to fly by it, updating the values of columns with each time step. The agent in Figure 1 below is black, nutrients in the world are green, and blue pixels indicate the agent’s field of view (what the organism can actually see around it).
While the agent always remains in the same column, it can execute one of three actions at any time based on what it sees in the environment to change its row. It can stay in its current row, or it can move upwards or downwards one row.
We would like to find agents who can feed themselves - that is, organisms that can observe nutrient locations and execute the appropriate actions to pass through those pixels and ingest them. We can judge how well an agent (a solution) is performing in this environment by enforcing some kind of an objective. Some hard objectives would be to eat every piece of food that passes through its FOV, or simply eat the most food during a lifetime. A softer objective might be to just eat enough food to stay alive based on some definition of its metabolic baseline.
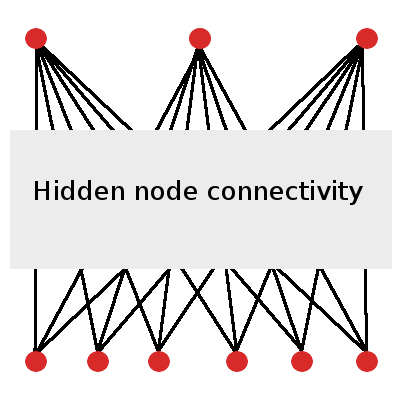
If we designed a neural network for this controller, it would require at least six input nodes (one for each of the pixels in its field of view) and three output nodes (one for each of the potential actions). An effective controller ANN may also include layers of hidden nodes. (Figure 2)
The evolearn package can be used to design this basic environment for one or even multiple agents to forage for nutrients in these pixelated “Grid World” environments. From this testbed we can compare the dynamics of populations of agents whose selection is restricted by the availability of resources at a given time, and represented by the encoding of the particular agent’s neural network in its genome. The encoding we choose to implement for a genome can in turn be restricted to those of more orthodox research paradigms, mainly direct encoding schemes where each characteristic of the network must be listed in its genome, and more contemporary approaches that attempt to compress features of an organism into smaller genomes, known as indirect encoding.
This compression provides a interesting testbed for introducing operators for mating and mutation to explore parameter space more efficiently than direct encoding schemes. Importantly, this approach has practical value to the study of biological organisms, as we come to appreciate the highly distributed and interdependent encoding of genetic information in DNA. Life has arrived at indirect encodings through natural selection in order to more greatly cover this space of possible organisms as a solution for solving endless objectives of metabolism and environmental pressures, and it is possible that the future of optimization engineering and artificial intelligence research may rely on mimicking these naturally-derived solutions.