Neuroevolution: A Primer¶
If we would like to evolve a population of our example agents on the task described, it is important that we have a stable representation of an artificial neural network, which we will use to represent that agent’s brain. With that representation, we can choose how to represent that network in a genome, making it possible to have two agent’s reproduce and mutate to find new agent solutions.
Within the field of neuroevolution, there are generally two different types of neural network representations we can choose as our agent genome: direct encodings and indirect encodings.
Direct Encodings¶
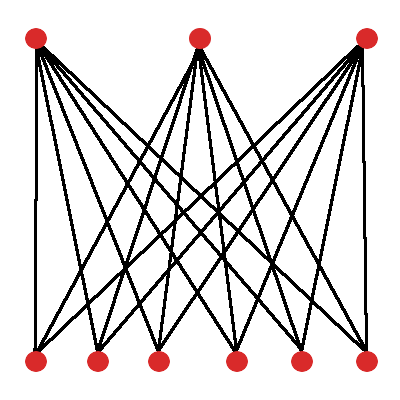
Since we need to find a representation for an agent in our population, let’s start with the simplest agent of that kind: a fully-connected neural network with 3 output nodes, 6 input nodes, and no hidden nodes. (Figure 3)
A directly encoded genome contains a one-to-one relationship between the parameters that describe the neural network and the number of genes in the associated agent genome.
For example, an agent with the network shown in Figure 3 has 18 unique weights, and therefore a direct encoding of that agent will require at least 18 values. It may additionally require information about the number of inputs, outputs, the activation functions used in those outputs, as well parameters that influence its learning within the environment.
For the purpose of our very simple task, 18 weight values is not very much. It would not be difficult to apply standard gradient-based learning to find the appropriate parameters for this model. If we did want to represent the network with a genome, 18 genes for each of the weights is relatively small, and we don’t run into many problems performing reproduction on mutation on it.
A direct encoding that will turn out to be essential to the approach of epann is known as NeuroEvolution of Augmenting Topologies, or NEAT. In the original paper that described the NEAT algorithm, a simple controller much like our example was described genetically as containing two separate lists to describe its neural network phenotype. (Figure 4)
A neural network is first described by its node genome (Figure 4(a)). The node genome is a list of each node in the agent’s neural network, along with some characteristics about each node. Nodes have a type, which distinguishes them as being an input, output or hidden node. Individual nodes also have unique activation functions, that determine the function applied to its summed inputs.
An agent also has a connection genome (Figure 4(b)). The connection genome is a list of each connection in the agent’s neural network, and it also has a list of attributes that describe them. They have an identification number (or, as will be called later, innovation numbers), a description of the nodes that start and end that connection (soon to be referred to as a connection’s out node and in node, respectively), and a weight. Connection genes also contain a final binary attribute called the enable bit, that determines whether or not a connection described in the connection genome will actually be expressed in the final agent phenotype.
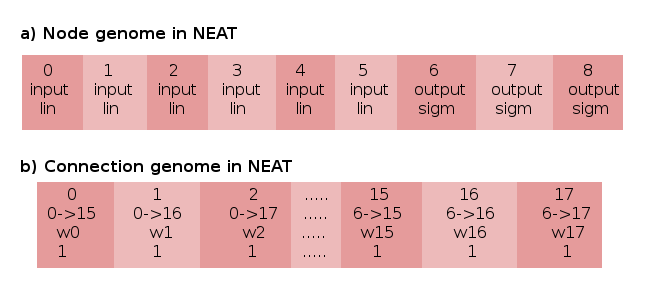
Direct Encoding in the original NEAT algorithm. Node and connection numbering maintain Python’s 0-based indexing left to right.
However, a direct encoding present difficulty as a general purpose representation for neural networks. Direct encodings will also require as many genes as there are parameters for more complex models, such as modern benchmark convolutional neural networks, which may contain over a million parameters. Using a genetic algorithm in a space that large will be difficult, and it is likely that candidate solutions will bounce around the space without converging.
Indirect Encodings¶
We do not see an analog to direct encoding in the genomes of biological organisms. Instead, organisms have genotypes with less information than the phenotypes they generate. The number of genes in a genome is far less than it would take to encode individual connections in its nervous system, let alone features of every cell in its body.
Instead, it seems that the emergence of life has settled on genetic representations that are far more complex. Genes are associated with the development of many different systems within an organism. As a result, a mutation in a particular gene can have widespread effects.
Indirect encodings can also change the search space we are exploring during our optimization.
In the field of neuroevolution, different indirect encoding frameworks have been proposed to accomodate potentially immense phenotype search spaces. It turns out, an effective indirect encoding can be implemented that is a variant of the NEAT direct encoding. The candidate indirect encoding is called Compositional Pattern Producing Networks (CPPNs) [CPPN2007], and they act as effective genome structures for a modified version of the NEAT algorithm: Hypercube-based NEAT. [ES2012]
| [CPPN2007] | Stanley, Kenneth O. “Compositional pattern producing networks: A novel abstraction of development.” Genetic programming and evolvable machines 8.2 (2007): 131-162.) |
| [ES2012] | Risi, Sebastian, and Kenneth O. Stanley. “An enhanced hypercube-based encoding for evolving the placement, density, and connectivity of neurons.” Artificial life 18.4 (2012): 331-363.) |